
Genomics is the study of whole genomes of organisms, and incorporates elements from genetics. Genomics uses a combination of recombinant DNA, DNA sequencing methods, and bioinformatics to sequence, assemble, and analyse the structure and function of genomes.
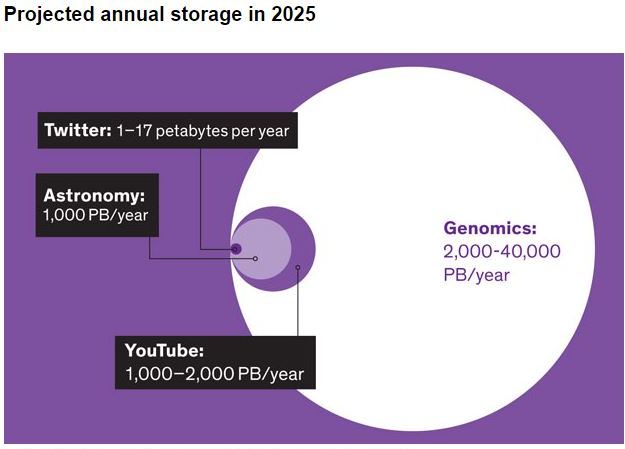
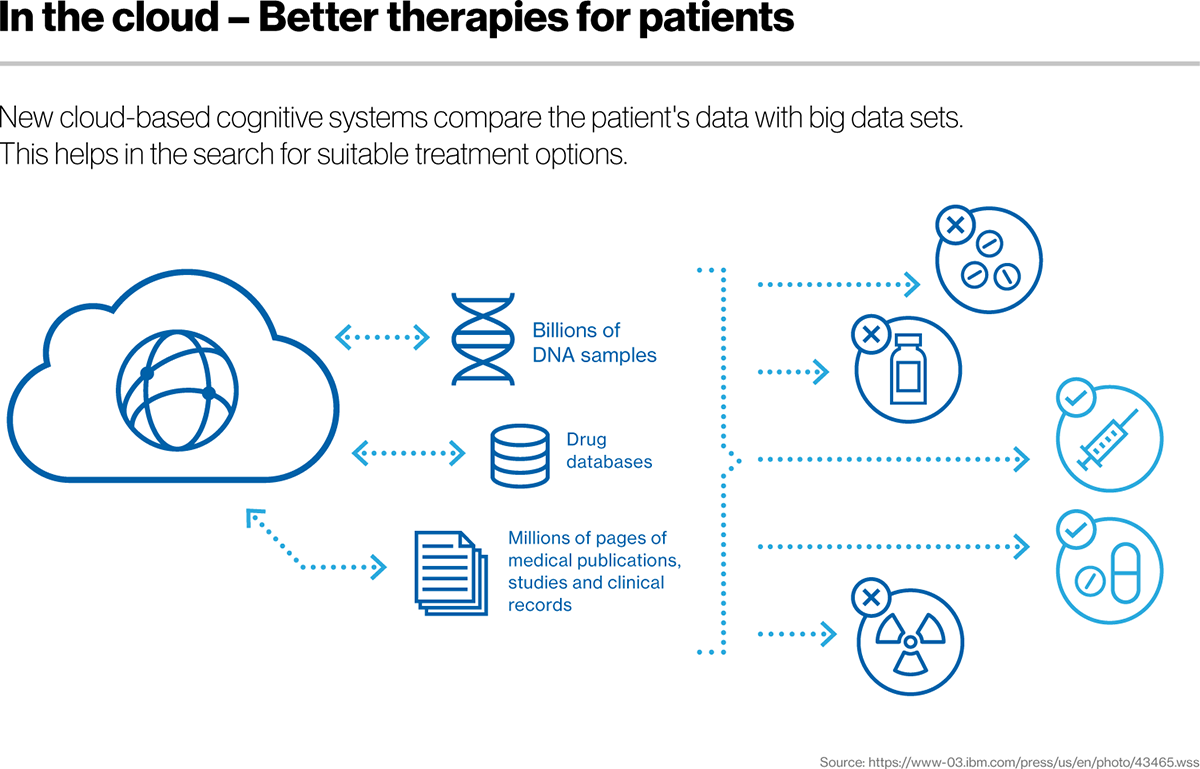
We are living in the time where big datasets are easily available and can be used for finding interesting research questions. Biological data in the form of genome sequencing and electronic health records can reveal novel traits which may help in improving life expectancy. Genome sequencing is becoming cheaper; therefore, it is now available even for developing countries and it is expected that prices will go down further. This would help clinicians to diagnose disease precisely that would open up to the area of precision medicine. Genome sequencing will also provide opportunity for the researchers to dig important biological information. There are several public databases such as ENCODE that provide TF binding (ChIP-seq), gene expression level (RNA-seq) and open chromatin region information (DHSs) for several cell types. There are also databases i.e., COSMIC and TCGA which contain cancer mutations. This Bioinformatics lab facility is being used to mine above public datasets and NGS (Next Generation Sequencing) data produced from local facilities for development of Predictors.
Abstract
Genomics research has become a global endeavor. The likelihood of identification of potential clinical variation increases with an increase in ability to delve deeper into human DNA. The Genomics lab aims to create a flexible, sustainable and competitive environment to support provision of prudent health-care services to the population of Pakistan. The main objective is to develop prediction methods for biological problems; based on machine learning and statistical methods which will subsequently increase the understanding of biology behind genetic abnormalities. This objective will bring many benefits for the patients including early diagnosis of disease, supporting better targeting therapies and quick recovery from disease. It also aims to train human resource for processing NGS (Next Generation Sequencing) data in order to make NGS common in Pakistan and to strengthen the national Genetic testing companies which will revolutionize public health by providing rapid, cost-effective and more accurate genetic tests.
Co-PI Details
Information
Name: Dr. Nisar Ahmed Shar
Designation: Assistant Professor
Department: Biomedical Engineering
Telephone: +92-21-99230604
Email: nisarshar@neduet.edu.pk
Research Interests
My research interests include development of prediction methods for biological problems based on machine learning and statistical methods. Recently, we have developed methods for prediction of transcription factors mutual interactions (Protein-Protein interactions) from ChIP-seq data generated by the ENCODE consortium, and did a conservation analysis of known functional Genomic regions. Realizing the importance of regulatory regions in controlling expression of genes, we have identified regulatory regions for 9000 genes that are mostly globally expressed genes and some of them are known to involve in cancer. Recently researchers have identified that regulatory regions harbour significant number of somatic mutations and our identified regulatory regions also contain high frequency of somatic mutations, which is an indication of positive selection.
Now researchers around the globe are realizing the importance of electronic health records, as they contain interesting information that can lead to the development of prognosis and diagnosis tools. As a data scientist, I am eager to utilize anonymized patient health records for development of prediction tools.
Genomics Lab Introduction
We are living in the time where big datasets are easily available and can be used for finding interesting research questions. Biological data in the form of genome sequencing and electronic health records can reveal novel traits which may help in improving life expectancy. Genome sequencing is becoming cheaper; therefore, it is now available even for developing countries and it is expected that prices will go down further. This would help clinicians to diagnose disease precisely that would open up to the area of precision medicine. Genome sequencing will also provide opportunity for the researchers to dig important biological information. There are several public databases such as ENCODE that provide TF binding (ChIP-seq), gene expression level (RNA-seq) and open chromatin region information (DHSs) for several cell types. There are also databases i.e., COSMIC and TCGA which contain cancer mutations.
This Bioinformatics lab facility is being used to mine above public datasets and NGS (Next Generation Sequencing) data produced from local facilities for development of Predictors.
Research Projects
1. Identification of gene specific cis-regulatory elements during differentiation of mouse embryonic stem cells: An integrative approach using high-throughput datasets
This study is focused on the mechanisms of gene-specific transcriptional control. by integrating gene expression data for the in vitro differentiation of murine ES cells to macrophages and cardiomyocytes, with dynamic data on chromatin structure, epigenetics and transcription factor binding. Combining a novel strategy to identify communities of related control elements with a penalized regression approach, we developed individual models to identify the potential control elements predictive of the expression of each gene. Our models were compared to an existing method and evaluated using the existing literature and new experimental data using embryonic stem cell differentiation reporter assays. Our method is able to identify transcriptional control elements in a gene specific manner that reflect known regulatory relationships and to generate useful hypotheses for further testing.
2. Identification of epigenetic markers for prediction of liver cancer
Liver cancer is the third most frequent causes of cancer related death accounting for more than 700,000 deaths each year. Most people die due to late diagnosis of disease. The identification of novel cancer causing genes and driver mutations provide targeted therapy for treating the disease. The present study aims to identify a set of novel genes and pathogenic alleles (consistent mutations) that might be involved in liver cancer. The non-coding mutations were identified in transcription factor binding sites of HepG2 cells. The pathogenic alleles identified in this study may help to understand the progression of liver cancer at molecular level. They may also act as potential biomarkers and therapeutic targets for liver cancer prediction and treatment.
Current Research Projects
1. Predicting the chance of occurrence of Liver Cancer in Hepatitis B & C patients
2. Development of predictors for early diagnosis of Breast Cancer
3. Selection of genes for Breast Cancer genetic test
4. Method development for prediction of Dengue outbreak in Pakistan
Recent Research Papers
1. Vijayabaskar, M., Goode, D. K., Obier, N., Lichtinger, M., Emmett, A. M., Abidin, F. Z., Shar, N., et al. (2019). Identification of gene specific cis-regulatory elements during differentiation of mouse embryonic stem cells: An integrative approach using high-throughput datasets.. PLoS computational biology, 15 (11), e1007337. https://doi.org/10.1371/journal.pcbi.1007337
2. Analysis of evolutionary patterns in the species of genus Homo
3. Repurposing drugs for the treatment of Epilepsy
4. Assessing the impact of mutations in viruses on disease spread
Collaborators
1. Jinnah Postgraduate Medical Centre Karachi
2. PCSIR Laboratories Karachi
3. Wellcome Sanger Institute Cambridge, UK


